Pancreatic Cancer: A Global Burden
Introduction
Cancer is an enormous burden on developed countries. Due to the growth and adoption of lifestyle behaviours such as poor diet, smoking and physical inactivity, the burden is projected to grow globally [1]. The GLOBOCAN 2018 data estimates that the rate of new cancer cases is reached 18.1 million while deaths related to cancer are about 9.6 million around the world. Approximately 2.5% of new cases occur because of pancreatic cancer PC and 4.5% death cases linked with pancreatic cancer [2]. PC statistics is confirmed that PC is the 12th ranked widely known type of cancer in men and the 11th ranked most common type of cancer in women throughout the globe. In 2012, 338,000 new cases were recognised and 4.1 in 100,000 people was found as a five-year average prevalence rate [3]. GLOBOCAN 2015 data was confirmed that 1000 PC cases are diagnosed daily throughout global [4]. Pancreatic cancer adenocarcinoma (PDAC) is an exocrine PC. PDAC is considered as the most common type of PC causing death [5]. Surgery is the ideal method of non-metastatic Pc treatment [6]. Although surgery has been used largely in local treatment, survival rates are modest due to PC metastasis. Radiotherapy and systemic therapies are utilised as therapeutic agents. Only 9% of PC is diagnosed in early stages, whereas the majority of cases present with distant metastasis to other organs which are reached 52% [7]. Many studies confirmed that Epithelial-to-Mesenchymal Transition (EMT) contributes largely to diverse carcinomas including PDAC [8]. EMT is identifiable with cell to cell loss adhesion and acquired motile mesenchymal characteristic which leads to cell invasion and migration. The moment migration cells reach their destination, they undergo a mesenchymal-epithelial transition (MET). Once MET starts, the secondary tumours establish and cause cancer metastasis and treatment failure [9]. The researcher has confirmed a relevance between metastasis and altered miRNA expression. For those seeking in-depth analysis, healthcare dissertation help can provide essential insights into the complexities of this disease.
MicroRNAs
MicroRNAs (RNAs) are minute (20-25 nucleotides) aligned in non-coding sequences and are detected in PC, particularly in early stages. MiRNAs triggers various diseases including PC. Recent studies have considered that miRNAs are both prognostic and predictive diseases markers. Indeed, miRNAs are being developed as therapies [10]. MiRNAs are expressed in the same way of any type of gene expression. MiRNA is crucial to comprehend how it contributes to disease and regulates biological pathways. MiRNA expression beings in the nuclease and is known as pre-miRNA. Drosha starts to remove the tail of primary RNA and leaves a short stem-loop structure deemed as pre-miRNA. This structure leaves the nucleus with exportin-5 association to the cytoplasm. In cytoplasm, the pre-miRNA is released from exportin-5 and starts a new process. Dicer complex result is an asymmetrical double-stranded miRNA by removing the stem-loop from the pre-miRNA. Mature miRNA is produced after duplex unwinding. It may integrate into a protein identified as RNA silencing complex (RISC). This complex from miRNA and RISC mediates gene silencing by cleaving and degradation mRNA or suppression of the translation process [11]. MiRNA is characterized with 3` untranslated regions (3`-UTR) of specific mRNA causing translation or degradation inhibition [12]. Particularly, these endogenous RNAs binding to (3`-UTR) of the complementary mRNA sequence, resulting in translation suppression and gene silencing. Approximately 30% of genes are controlled by miRNAs. Studies have demonstrated that miRNAs are regulated thousands of human protein-coding genes. Currently, understanding of miRNAs is indicated that miRNAs have a broader impact on both the evolution and expression of protein-coding genes [13]. The significance of miRNAs in cancer biology has been underlined by the identification of alterations in miRNAs processing machinery in cancer cell [14]. Various studies have demonstrated the major role of miRNAs in cancer biology through regulating the mRNAs target expression to stimulate tumour growth, angiogenesis, invasion and immunity evasion [15]. They contribute significantly to regulatory role in the differentiation, development and apoptosis of the normal cell. Moreover, they determine the final phenotype of cancers cells and metastatic potential [16].

MicroRNA 21
MicroRNA 21 (miRNA 21) contributes largely to upregulated miRNAs in solid tumours. It was identified as a first mammalian conserved across vertebrate diverse species evolution [17]. However, the location is different between humans and other vertebrate species. In humans, miRNA 21 is located on chromosome 17q23.2. miRNA 21 is located in intronic regions of a protein-coding gene where the primary transcript of the miRNA 21 gene is independently transcribed from its promoter regions [18]. Experimental data have demonstrated that in many cells’ types, miRNA 21 function is a pro-survival and anti-apoptotic factor [19]. These data have confirmed that miRNA 21 high levels of expression may lead to oncogenic effects, whereas the low level of miRNA 21 expression temporarily required for differentiation and development [20]. MiRNA 21 is a trigger in wide diverse types of cancers including breast, colon, lung, prostates stomach cancer and pancreatic cancer. It is considered as a typical (onco-miRNA) because it inhibits phosphatase expression thus limiting signalling pathway activity such as AKT signalling [21]. It has been identified in pre-B-cell lymphoma and inhibition of miRNA21 cause to induce the biological alternations in diffuse large B-Cell lymphoma (DLBCL) which is linked with AKT signalling pathway that is activated during carcinogenesis [22]. MiRNA 21 has been investigated as one of gemcitabine resistance. It has a significant associated with short survival time. The researcher suggested that miRNA 21 contributes to gemcitabine chemoresistance via phosphatase and tensin homolog (PTEN) an inhibiting tumour suppressor gene that activates the AKT pathway. PTEN is a gene which corresponding with many different of mammals’ cancer. PC is one of them. PTEN gene does many biological processes. It is capable of causing negative regulation of phosphatidylinositol 3-kinase (PI3K) signalling. PI3K is a mediated factor to any processes that prevent or extent of signal transduction. It also acts as a tumour suppressor gene via activating of its phosphatase protein product. This action leads to inhibit the AKT signalling pathway which plays as a key role of regulating cellular behaviours for instant cell migration and cell growth [23].
MicroRNA 155
MicroRNA 155 (miRNA 155) is one of the most important up-regulation miRNAs in solid tumour. It is a well-conserved sequence in mammals. It plays a major role in various pathological and physiological processes. MiRNA 155 is considered as the first line of innate immune system defence against invading pathogens. The overexpression of miRNA 155 causes decreased levels of tumour protein-53-induced- nuclear-Protein 1 (TP53INP1) in PC. Whereas, the over-silencing by miRNA 155 leads to activated oncogenic cascades which are triggered by apoptotic resistance and subsequently silences pro-apoptotic TP53INP1 [24]. Furthermore, the downregulation of tumour protein 53 (TP53) induces cell growth apoptosis and inhibition by altering TP53 transcriptional activity. This is linked with increase of miRNA 155 in PC [25]. MiRNA 155 in PC cells significantly contributes in cancer cell invasion and migration regulation by reducing the SOCS1 expression and modulating the STAT3 signalling pathway. Suppressor Cytokine signalling 1 (SOCS1) is deemed as a gene that suppresses tumour .It is normally a negative feedback regulation for Janus activated Kinase (JAK)/signal transducer and activator of transcription-3 (STAT3) signalling. As EMT is regulated via a complex signalling network. STAT3 signalling pathway was proved to be involved in pancreatic cancer EMT [26]. Cancer-associated fibroblasts (CAF) are the PC microenvironment major constituents. PC cells are capable of inducing normal fibroblasts (NF) converting to CAF subsequently promoting tumour proliferation and invasions. Microvesicles (MV) have confirmed to be intracellular communication major mediator and they can be involved in transportation of secret miRNA to the recipient cell from the donor cell. Studies have proved that miRNA 155 was up-regulated in PC donor MV and the NF has converted into CAF after absorption of MV containing miR155. In addition, the TP53INP1 protein levels down regulation contributes to fibroblasts activation because TP53INP1 is a miRNA 155 target in fibroblasts [27]. MiRNA 155 expression has demonstrated to stimulate gemcitabine to lead to increased miRNA 155 expression and that increases exosome production and inhibits apoptosis, resulting in gemcitabine resistance [28].
Methods:
In Situ Hybridisations (ISH) was executed to determine miRNA 21 and miRNA 155 levels in both normal pancreas tissue and pancreatic cancer tissue. ISH was applied in the University of Westminster, Cavendish Campus Laboratories, London, United Kingdom. ISH assay was used to localise molecular information in the specific tissues. This assay requires evaluating the target signalling within a morphology context. Therefore, this assay requires an accurate between the preservation of a recognisable constructional framework and full availability of hybridisation target [29]. ISH is usually performed by designing on antisense probe to the main target mRNA which makes the probe and miRNA to bind and then visualizing in the tissue slide. Human pancreas and adjacent normal pancreas tissues were purchased from Super Biochips laboratories. Additional patient slides from The Liver Institute, King's College London were used. MTA was applied for this. Thus, no ethical approval was required. All slides were formalin-fixed paraffin-embedded (FFPE) tissue. FFPE samples had been chosen because they are the most common tissue for achieving tumour samples and can be stored at room temperatures. They also work well for morphology analysis. FFPE sections size were 5μm. All sections were labelled as a case number 1,2 and 3. Each case had many regions to detect from normal and cancer area which was labelled as A, B and C. ISH was performed for each case individually. After labelling slides, all sections were prepared for ISH by dewaxing.
Dewaxing
Xylene was used two times for 3 min. each. Following with ethanol 99% once for 4 min. Then sections were cleansed with diethylpyrocarbonate (DEPC) treat water for 5 min. After that, all slides were placed on Omnislide. Omnislide had already set up at 37°C and some wet tissues were put in the margin to provide a humidity environment and prevent slides from drying. Sufficient amount of Proteinase k (PK) was added on each slide to completely cover the sections. Two concentrations of PK were performed. First, concentration was 5μg/ml. It was made by adding 975 μl of Tris-HCl with 25μl of PK in a new eppendrof. Second concentration was 2μg/ml. It was made by adding 990 μl of Tris-HCl with 10μl of PK in a new eppendorf. Both concentrations were applied in case 1C to optimize tissue permeabilization. Dewaxing steps were finished by washing two times with DEPC for 5min. each. The solvent xylene is an organic solvent which is used to remove any remaining paraffin from the slides and make them transparent because paraffin can fully envelop tissue. Due to the fact that xylene is immiscible with water, ethanol 99% was used as a "water-free" than washing with DEPC treated water. DEPC treated water inactivates RNase enzymes in the water. It is used in the laboratory to reduce the risk of RNA degradation by RNase. These steps were performed to unmask latent epitopes which make the antibodies completely access to antigens in the tissue and bind to the correct epitopes. Treatment with PK is an important step to make access to the target nucleic acid. It was used to open the cell membranes. PK digestion is a critical step in ISH assay. The insufficient digestion leads to diminishing hybridisation signalling. Nevertheless, the over digestion lead to damage the tissue morphology and make the hybridisation signal impossible to localise. Many factors might optimal PK concentration such as the size of the tissue, length of fixation and tissue type. This experiment was applied 2-5 μg/ml PK for 30min.
Hybridisation
All slides were dried from DEPC and replaced on Omnislide. Then, 50μl of prehybridisation was applied on each slide and incubate for 1hr. at 37°C. Following the step, an equal volume of miRNA probe (dig-labelled miR21, miR 155 and scrambled) was applied on sections and incubated at 37°C overnight. Next day, hybridisation was finished by washing the slides with Tris-buffer-saline (TBS) two times for 5 min. each. Prehybridisation means before hybridisation. The main purpose of this step to reduce the amount of probe required and the hybridisation time by using sufficient hybridisation buffer to cover the sections. It is considered as a hybridisation incubation without the labelled probe which prevents the probe to bind unspecific site instead of the target. After that, three types of probes were applied. All these probes were digoxigenin (non-radioactive) labelling system. Dig labelling was used to support post hybridisation detection by the formation of an insoluble colour participate directly on a filter membrane. In ISH assay Dig Oligonucleotide labelling probe is the best hybridisation probe because it is designed by using anti-dig antibody. MiRNA 21and miRNA 155 probes are antisense probe to detect the target miRNA 21 and miRNA 155 in the tissue. The scrambled probe was used as a negative control because it does not identify any miRNA sequences in FFBE human tissue. It was used to optimize the sensitivity and the performance of ISH because it does not have any specific target. TBS was used to maintain the pH with a narrow range. It was also used to wash because its chemical structure does not antibody – antigen binding interaction. TBS was prepared by measuring 6.05 g/l of Tris-(hydroxymethyl) aminomethane in weighing boat and was poured into 1L bottle to make 50 mM Tris. 8.766 g of NaCl was measured and added in the same bottle. Then, 1L of D.W was added. Finishing preparing steps by the adjusting pH to 7.6.
Detection
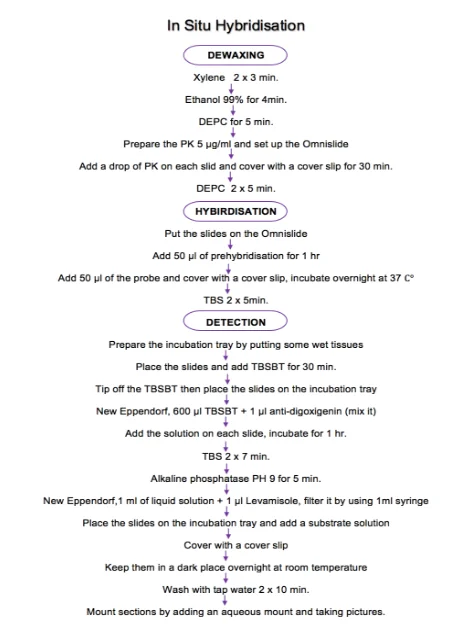
After washing with TBS, all the slides were placed on the incubation tray, which had already put some wet tissues in the margin. An equal amount of TBSBT was added on each slide. After 30min., all slides were tipped off and replaced on the incubation tray. New Eppendorf was performed to mix 600 μl TBSBT with 1 ml anti-digoxigenin alkaline phosphatase antibody (ROCH). Sufficient amount of the solution was added on each slide for 1 hr. Then all the slides were cleansed with TBS two times for 7 min. each. After washing all slides were immersed in alkaline phosphatase pH9 for 5 min. Following step, New Eppendorf was prepared to add 1μl of liquid solution Janus activated Kinase (JAK) substrate solution (Sigma Aldrich, UK) with 1mM of Levamisole. The NBT/BCIP visualization generates a blue precipitate detectable by using microscopy light. Slides were placed on the incubation tray and a sufficient amount of substrate solution was added. All slides were incubated in the dark place overnight after covering them with a cover slip. Following day, all slides were washed with tap water two times for 10 min. Finally, all sections were mounted by adding an aqueous mount. All pictures were taken by using a microscope. TBSBT was used to dilute anti-digoxigenin. TBSBT was consist of (TBS containing 0.1 % levels of Triton-X-100) and approximate 3% levels of bovine serum albumin. Alkaline phosphatase was used in this stage of ISH because it is dramatically increased the ISH signal with these probes. It was used to detect the target miRNA. Anti-digoxigenin alkaline phosphatase conjectures antibody was used as a secondary antibody. NBT/BBCIP solution was used to detect the alkaline phosphatase because of the solution sensitivity. Levamisole was used to reduced background alkaline phosphatase activity in ISH assay and increase the detection of signalling target. All slides were incubated in the dark place because the solution has a light sensitivity. Finally, the aqueous mount was used to preserved and support sections from drying. All results were collected by taking pictures from the microscope which connected to the computer. CytoCam was the program which read all the results. All pictures were taken by switching off the white balance to get better resolution. Diagram (1) shows all the steps of ISH.
Results:
The ISH was performed on each case individually. Various areas from each case were examined. These areas related to infection and non-infection tissues from the same patient. All cases had already diagnosed with PDAC. Some cases were suffered from the tumour mass and others from the lesion. ISH was applied in each case to detect the level of miRNA 21 and miRNA 155 in this case. Sections were collected from normal and abnormal tissues from each case. ISH was performed on patient 2005 were evaluated with a PDAC tissues samples analysed. All cases were labelled as shown below in details.
Pancreatic Ductal Adenocarcinoma Cases (1,2 and 3 from 2005)
Case 1
1B = intrapancreatic mass, 1C= Resection Margin of Pancreas.
Case 2
2A= Tumour mass, 2B= Pancreatic Background.
Case 3
3B= Pancreatic Parenchyma, 3C= Lesion.
Case 1 (1B and 1C)
Case 1 was a patient who was applied for biopsy. Case 1 tissues were taken from the intrapancreatic mass area and resection margin of the pancreas. ISH was performed on case 1. Different concentration of PK was used in 1C sample in order to confirm the overexpression of miRNA 21 and miRNA 155 and optimize the results. Both concentrations of PK in 1C sample are shown a significant level of staining. Furthermore, the abnormal cells are shown clearly in the resection margin area. Fig.1 illustrates the higher expression of miRNA appearing in the second and third column 1C PK2,1C PK5 respectively. Both columns have the highest staining with miRNA 21 and miRNA 155 rows compared with the scrambled row which does not have any level of staining. By contrast, the negative staining of miRNA is shown in the intrapancreatic mass which illustrated in the 1B PK2 column from Fig.1. The normal cell morphology in this area is correlated with negative staining. The level of staining has not changed in miRNA21, miRNA 155 and scrambled in the intrapancreatic mass area. All samples have not been shown any changing of staining in the scrambled row.
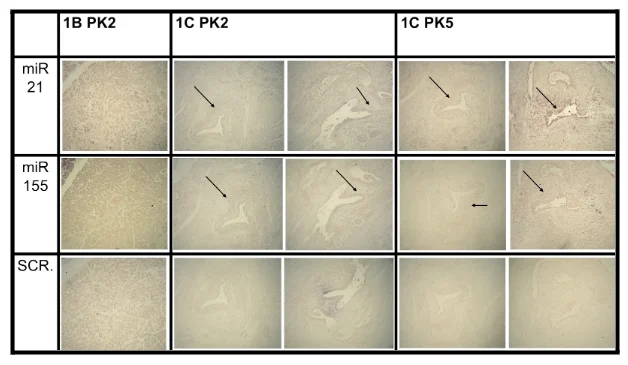
Figure 1 miRNAs expression in human pancreatic cancer tissues and adjacent normal pancreas tissues. In Situ Hybridisation analysis for miRNAs. First row shows the expression of miRNA 21 in each case. Black arrows demonstrated the high expression areas (positive staining). Second row shows the expression of miRNA 155 in each case. Black arrows demonstrated the high expression areas (positive staining). Third row shows the miRNA scrambled (scr.) results which used as a control. Where 1B = intrapancreatic mass, 1C= Resection Margin of Pancreas PK2=proteinase K 2μg/ml, PK5= proteinase K 5μg/ml
Case 2 (2A and 2B)
Case 2 was a patient who applied for biopsy. Case 2 tissues were taken from the tumour mass and pancreatic background. ISH was performed on case 2. Both samples, 2A and 2B were treated with 2μg/ml from proteinase K. The high level of miRNA 21 and miRNA 155 expression are shown in the tumour mass area. Moreover, the abnormal cells are shown obviously in this tumour mass area. Fig.2 illustrates miRNA 21 and miRNA 155 high levels of expression appearing in the first column. 2A PK2 column has the highest expression miRNA 21 and miRNA 155 rows compared with the last row (scrambled row) which does not demonstrate any level of staining. By contrast, the pancreatic cancer background area is shown negative staining which illustrates in the second column from Fig.2. The morphology of cells in pancreatic background area was normal and it is correlated with negative staining. The level of staining in the pancreatic background has not changed with miRNA 21, miRNA 155 and scrambled. Both samples 2A and 2B from case 2 have not changed in a scrambled row.

Figure 2 miRNAs expression in human pancreatic cancer tissues and adjacent normal pancreas tissues. In Situ Hybridisation analysis for miRNAs. First row shows the expression of miRNA 21 in each case. Black arrows demonstrated the high expression areas (positive staining). Second row shows the expression of miRNA 155 in each case. Black arrows demonstrated the high expression areas (positive staining). Third row shows the miRNA scrambled (scr.) results which used as a control.
Where 2A= Tumour mass, 2B= Pancreatic Background
PK2=proteinase K 2μg/ml
Case 3 (3B and 3C)
Case 3 was a patient who was applied for biopsy. Case 3 tissues were taken from the pancreatic parenchyma and lesion area. ISH was performed on case 3. Both samples 3B and 3C were treated with 2μg/ml of proteinase K. Both of them have not shown any staining neither miRNA21 nor miRNA 155 as Fig.3 shows. Both of them are shown the same level of staining in each row miRNA 21, miRNA 155 and scrambled. The morphology of the cell was normal in each sample correlated with negative staining.

Figure 3 miRNAs expression in human pancreatic cancer tissues and adjacent normal pancreas tissues. In Situ Hybridisation analysis for miRNAs. First row shows the expression of miRNA 21 in each case. Second row shows the expression of miRNA 155 in each case. Third row shows the miRNA scrambled (scr.) results which used as a control. Where 3B= Pancreatic Parenchyma, 3C= Lesion
PK2=proteinase K 2μg/ml.
Taking together, results are collected and shown in Fig.4. The abnormal morphology has appeared in 1C PK2, 1C PK5 and 2A PK2 samples correlated with positive staining with miRNA 21 and miRNA 155. By contrast, the normal phenotype cells have appeared in 1B PK2, 2B PK2, 3B PK2 and 3B PK2 with negative staining in miRNA 21 and miRNA 155. All samples are shown negative staining with scrambled which confirmed the performance optimization and sensitivity. Table 1 is shown all the results.

Figure 4 miRNAs expression in human pancreatic cancer tissues and adjacent normal pancreas tissues. In Situ Hybridisation analysis for miRNAs. First row shows the expression of miRNA 21 in each case. Black arrows demonstrated the high expression areas (positive staining). Second row shows the expression of miRNA 155 in each case. Black arrows demonstrated the high expression areas (positive staining). Third row shows the miRNA scrambled (scr.) results which used as a control.
Where 1B = intrapancreatic mass, 1C= Resection Margin of Pancreas,2A= Tumour mass, 2B= Pancreatic Background, 3B= Pancreatic Parenchyma, 3C= Lesion.
PK2=proteinase K 2μg/ml, PK5= proteinase K 5μg/ml.

Discussion:
The results indicates that high levels of miRNA 21 and miRNA 155 expression can endow pancreatic cancer cells with two aspects. First, promoting migrates of tumour cells and keeps them survival. Second, increasing resistance to chemotherapy. First, the promoting migrates of tumour cells provided a survival mechanism for pancreatic cancer cells and then metastasis happens. Metastasis occurs when cell loss cell to cell contact. Over these years, researcher has proved that EMT contributes largely in cancer metastasis and chemoresistance. EMT triggers diverse regulatory mechanisms. Particularly, the PI3K/AKT signalling pathway stimulates EMT in diverse ways [9]. The researcher demonstrated that miRNA contributes significantly in cancer progression by inactivating tumour suppressor genes and activation the oncogenes [13]. Identification of alternations in miRNAs has been considered as a prognostic marker in cancer biology [14]. Moreover, the final phenotype of cancer metastasis has been determined by miRNAs [16]. MiRNA 21 is known also as upregulated pathways in human malignancy including PC. It suppresses a large number of gene expression [21]. One of the miRNA21 targets is PTEN gene. PTEN gene is described as tumour suppressor gene which inhibits the AKT pathway by negating PI3K regulation. The PI3K/AKT signalling pathway affects the EMT in diverse ways which influences tumour aggressiveness [19]. MiRNA 155 is one of the upregulation miRNAs in solid tumour. The overexpression of miRNA 155 in PC leads to downregulation of SOCS1. SOCS1 downregulation causes the negative effect of STAT3 signalling pathway which is led to induce EMT cancer cell in PC [26]. Furthermore, the overexpression of miRNA 155 leads to repressing the TP3INP1 expression which is considered as a proapoptotic stress-induced P53 target gene in PDAC. TP53INP1 is also a miRNA 155 target in fibroblasts and TP53INP1 protein down regulation converting the NF into CAF in PC [27]. This study suggested the miRNA 21 and miRNA 155 high regulations play a vital role in pancreatic cancer metastasis by re-regulating the EMT. EMT is considered as a first step to making treatment fail and metastasis happens. Second, the increasing resistance to chemotherapy, Recent studies have demonstrated that chemotherapeutic resistance/sensitivity is related to specific miRNA expression in PC and regulation of molecular signalling pathway [5].
MiRNA 21 is investigated as a chemotherapeutic treatment resistance in PDAC especially with gemcitabine. Researchers confirmed that miRNA 21 participates with gemcitabine chemoresistance by inhibiting PTEN tumour suppressor gene via activation the PI3K/AKT signalling pathway [23]. MiRNA 155 was demonstrated as a gemcitabine resistance stimulator. Studies confirmed that the long period of gemcitabine leads to increase in inhabit the apoptosis and increases miRNA 155 expression .That means miRNA 155 plays contribute largely in chemotherapy treatment. This study suggested the high levels of miRNA 21 and miRNA 155 expressions play a significant role in chemotherapeutic resistance. This finding may throw light on a novel therapy to increase pancreatic cancer patient’s survival. The positive results in the first row of Fig.4 have confirmed the overexpression of miRNA 21 in PDAC. The overexpression is clear evidence that a link between miRNA 21 expressions and metastasis exists. The link is latent, as miRNA 21 is a tumour suppressor target and PTEN is the main target of miRNA 21 in PC. The overexpression of miRNA 21 leads to the negative regulation expression of PTEN. PTEN plays an important role in the inhibition AKT pathway. The inhibition of the AKT pathway cause PI3K negative regulation. More importantly, EMT is affected by the PI3K/AKT signalling pathway. The mis-regulation of EMT cells supports cancer cell migration. Once cancer cells start invasion to its destination, metastasis happens. Furthermore, based on fact, miRNA 21 overexpression in PDAC drives to increase the sensitivity of gemcitabine chemoresistance; miRNA 21 contributes greatly in tumour therapy treatment. The positive results have confirmed the overexpression of miRNA 155 in PDAC in the second row of Fig 4. The high level of miRNA 155 expression is given clear evidence that is a link between metastasis and miRNA 155 expression. The upregulation of miRNA 155 in PDAC let to decrease SOSC1 tumour suppressor gene expression. SOSC1 can modulate STAT3 signalling. STAT3 signalling has an important effect on EMT. It is considered as one of the complex signalling networks. As mentioned before, TP53INP1 is the miRNA 155 target in PDAC. The miRNA 155 higher expression leads to decreased expression of TP53INP1 protein. This downregulation reduced cell tumour suppressor and TP53INP1 is one of them. This downregulation of TP53INP1 also converts the NF to CAF which promotes tumour invasion and migration. MiRNA 155 in the high level of expression is also considered as a chemotherapy resistance by stimulating gemcitabine resistance. Overall, this study shows that miRNA 21 and miRNA 155 levels of expression are elevated in human pancreas tumours. In addition, it illustrated that miRNA 21 and miRNA 155 are necessary for the EMT induction in the pancreatic cancer cells by remodelling the signalling pathway. Finally, it shows that miRNAs are necessary for pancreatic cancer cell survival. Collectively; these observations suggest that miRNAs are considered as a probable therapeutic target for the novel approach treatment of pancreas cancer.

Acknowledgements
First, I would like to express deepest appreciation to my supervisor Dr. Pinar Uysal-Onganer, for her patience and consistent encouragement I received throughout the research work. Without her supervision, this thesis would have been implausible. Dr Tony Warford thank you for your pathological support that made this research work possible. Many thanks for all the people who work in the library for their help. My sincere gratitude to the lab technician staff who supported me throughout the research work.
References:
Torre, L., Bray, F., Siegel, R., Ferlay, J., Lortet-Tieulent, J. and Jemal, A. (2015). Global cancer statistics, 2012. CA: A Cancer Journal for Clinicians, 65(2), pp.87-108.
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R., Torre, L. and Jemal, A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians, 68(6), pp.394-424.
Tarver, T. (2012). Cancer Facts & Figures 2012. American Cancer Society (ACS). Journal of Consumer Health On the Internet, 16(3), pp.366-367.
Gibberd, R. (2000). Globocan 1: Cancer Incidence and Mortality Worldwide. J. Ferlay, D.M. Parkin and P. Pisani, IARC Press, Lyon, 1999. Price: $90. Statistics in Medicine, 19(19), pp.2714-2715.
Madurantakam Royam, Ramesh, Shanker, Sabarimurugan, Kumarasamy, Ramesh, Gothandam, Baxi, Gupta, Krishnan and Jayaraj (2019). miRNA Predictors of Pancreatic Cancer Chemotherapeutic Response: A Systematic Review and Meta-Analysis. Cancers, 11(7), p.900.
Hartwig, W., Hackert, T., Hinz, U., Gluth, A., Bergmann, F., Strobel, O., Büchler, M. and Werner, J. (2011). Pancreatic Cancer Surgery in the New Millennium. Annals of Surgery, 254(2), pp.311-319.
Jiang, J., Liu, C., Cheng, H., Lu, Y., Qin, Y., Xu, Y., Xu, J., Long, J., Liu, L., Ni, Q. and Yu, X. (2015). Epithelial–mesenchymal transition in pancreatic cancer: Is it a clinically significant factor?. Biochimica et Biophysica Acta (BBA) - Reviews on Cancer, 1855(1), pp.43-49.
Dart, D., Uysal-Onganer, P. and Jiang, W. (2017). Prostate-specific PTen deletion in mice activates inflammatory microRNA expression pathways in the epithelium early in hyperplasia development. Oncogenesis, 6(12).
Abreu, F., Liu, X. and Tsongalis, G. (2017). miRNA analysis in pancreatic cancer: the Dartmouth experience. Clinical Chemistry and Laboratory Medicine (CCLM), 55(5).
Kasinski, A. and Slack, F. (2011). MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nature Reviews Cancer, 11(12), pp.849-864.
Kim, V. (2005). MicroRNA biogenesis: coordinated cropping and dicing. Nature Reviews Molecular Cell Biology, 6(5), pp.376-385.
Selcuklu, S., Donoghue, M. and Spillane, C. (2009). miR-21as a key regulator of oncogenic processes. Biochemical Society Transactions, 37(4), pp.918-925.
Roldo, C., Missiaglia, E., Hagan, J., Falconi, M., Capelli, P., Bersani, S., Calin, G., Volinia, S., Liu, C., Scarpa, A. and Croce, C. (2006). MicroRNA Expression Abnormalities in Pancreatic Endocrine and Acinar Tumors Are Associated with Distinctive Pathologic Features and Clinical Behavior. Journal of Clinical Oncology, 24(29), pp.4677-4684.
Landgraf, P., Rusu, M., Sheridan, R., Sewer, A., Iovino, N., Aravin, A., Pfeffer, S., Rice, A., Kamphorst, A., Landthaler, M., Lin, C., Socci, N., Hermida, L., Fulci, V., Chiaretti, S., Foà, R., Schliwka, J., Fuchs, U., Novosel, A., Müller, R., Schermer, B., Bissels, U., Inman, J., Phan, Q., Chien, M., Weir, D., Choksi, R., De Vita, G., Frezzetti, D., Trompeter, H., Hornung, V., Teng, G., Hartmann, G., Palkovits, M., Di Lauro, R., Wernet, P., Macino, G., Rogler, C., Nagle, J., Ju, J., Papavasiliou, F., Benzing, T., Lichter, P., Tam, W., Brownstein, M., Bosio, A., Borkhardt, A., Russo, J., Sander, C., Zavolan, M. and Tuschl, T. (2007). A Mammalian microRNA Expression Atlas Based on Small RNA Library Sequencing. Cell, 129(7), pp.1401-1414.
Volinia, S., Calin, G., Liu, C., Ambs, S., Cimmino, A., Petrocca, F., Visone, R., Iorio, M., Roldo, C., Ferracin, M., Prueitt, R., Yanaihara, N., Lanza, G., Scarpa, A., Vecchione, A., Negrini, M., Harris, C. and Croce, C. (2006). A microRNA expression signature of human solid tumors defines cancer gene targets. Proceedings of the National Academy of Sciences, 103(7), pp.2257-2261.
Davis, R., Ngo, V., Lenz, G., Tolar, P., Young, R., Romesser, P., Kohlhammer, H., Lamy, L., Zhao, H., Yang, Y., Xu, W., Shaffer, A., Wright, G., Xiao, W., Powell, J., Jiang, J., Thomas, C., Rosenwald, A., Ott, G., Muller-Hermelink, H., Gascoyne, R., Connors, J., Johnson, N., Rimsza, L., Campo, E., Jaffe, E., Wilson, W., Delabie, J., Smeland, E., Fisher, R., Braziel, R., Tubbs, R., Cook, J., Weisenburger, D., Chan, W., Pierce, S. and Staudt, L. (2010). Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature, 463(7277), pp.88-92.
Giovannetti, E., Funel, N., Peters, G., Del Chiaro, M., Erozenci, L., Vasile, E., Leon, L., Pollina, L., Groen, A., Falcone, A., Danesi, R., Campani, D., Verheul, H. and Boggi, U. (2010). MicroRNA-21 in Pancreatic Cancer: Correlation with Clinical Outcome and Pharmacologic Aspects Underlying Its Role in the Modulation of Gemcitabine Activity. Cancer Research, 70(11), pp.4528-4538.
Gironella, M., Seux, M., Xie, M., Cano, C., Tomasini, R., Gommeaux, J., Garcia, S., Nowak, J., Yeung, M., Jeang, K., Chaix, A., Fazli, L., Motoo, Y., Wang, Q., Rocchi, P., Russo, A., Gleave, M., Dagorn, J., Iovanna, J., Carrier, A., Pebusque, M. and Dusetti, N. (2007). Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proceedings of the National Academy of Sciences, 104(41), pp.16170-16175.
Tam, W., Ben-Yehuda, D. and Hayward, W. (1997). bic, a novel gene activated by proviral insertions in avian leukosis virus-induced lymphomas, is likely to function through its noncoding RNA. Molecular and Cellular Biology, 17(3), pp.1490-1502.
Mikamori, M., Yamada, D., Eguchi, H., Hasegawa, S., Kishimoto, T., Tomimaru, Y., Asaoka, T., Noda, T., Wada, H., Kawamoto, K., Gotoh, K., Takeda, Y., Tanemura, M., Mori, M. and Doki, Y. (2017). MicroRNA-155 Controls Exosome Synthesis and Promotes Gemcitabine Resistance in Pancreatic Ductal Adenocarcinoma. Scientific Reports, 7(1).
Pang, W., Su, J., Wang, Y., Feng, H., Dai, X., Yuan, Y., Chen, X. and Yao, W. (2015). Pancreatic cancer-secreted miR-155 implicates in the conversion from normal fibroblasts to cancer-associated fibroblasts. Cancer Science, 106(10), pp.1362-1369.
Hankin, R. (1992). In Situ Hybridization: Principles and Applications. Laboratory Medicine, 23(11), pp.764-770.
- 24/7 Customer Support
- 100% Customer Satisfaction
- No Privacy Violation
- Quick Services
- Subject Experts



